
Louis Leakey was a Kenyan paleoanthropologist who devoted his life to uncovering the early phases of human evolution. Using the scientific tools available at the time, he set out to study hominid fossils in Olduvai Gorge, a ravine in the Great Rift Valley in eastern Africa. His discoveries proved that humans originally evolved in Africa, a hypothesis initially proposed by Darwin that had previously been met with skepticism by the scientific community.
Studying fossils is the traditional way of understanding how life forms on earth evolved over time. Fossils provide snapshots of ancient plants, animals, and microorganisms, and through comparison with current life forms as well as other fossil records, the gradual evolution of one species into another can be reconstructed. In addition, estimates of the ages of fossils often allow researchers to deduce how long ago a new species emerged, and the time when the last shared ancestor of two currently existing species roamed this planet. The evolutionary relationships between species are often represented in so-called phylogenetic trees or “family trees”. In a phylogenetic tree, each juncture represents the last common ancestor shared between the species at the tips of the branches.
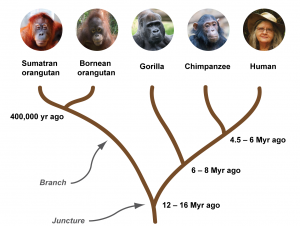
As we can see from the phylogenetic tree of the great ape family, orangutans were the first ones to split off from the great ape family tree and evolve into an independent species approximately 14 million years ago (Mya) [1]. Gorillas diverged from a common ancestor with chimpanzees and humans ~7 Mya, while the chimpanzee and human lineages separated around 5 Mya [1]. Historically, family trees were constructed based on information deduced from fossil records, and by comparison of the morphology (body part characteristics) of existing species. Morphology includes both the outer appearance as well as inner structures of living organisms. For example, if you compare the skeletons of a horse, an orangutan, and a human, you can easily see that the overall organization of their bones is similar – all three have four limbs, their lungs are encased by chest bones, and their jaws contain teeth. At the same time, it is also evident that orangutans and humans share more similarities than either species shares with the horse, which means that orangutans and humans are more closely related.
Although constructing phylogenetic trees from fossil records is a powerful method, there are significant limitations: research is limited to species that were preserved well over millions of years, and to fossils that can be or have been located. Therefore, fossil records often leave researchers with a “missing link” problem: they know that an intermediate form must have existed, bridging the gap in development between two known fossils or a fossil and a current species, but this “missing link” has yet to be found. Also, important information about the biology of a species, such as their behavior, their preferences for certain food, and their interactions with each other, remain elusive as it is difficult to extract this “soft” information from bones.
To overcome the limitations of studying fossils to understand human evolution, Louis Leakey decided to complement his archaeological “bones and stones” research in Africa with field studies of the great apes, our closest living relatives, in their natural habitats [2]. His quest was to unravel the characteristics of the last common ancestor we shared with chimpanzees, gorillas, and orangutans. He also hoped to gain insights into environmental and other factors that triggered the evolution of humans [2]. He assumed that the environment in which early humans evolved was similar to the current habitat of chimpanzees and gorillas in Africa, and he therefore focused his efforts initially on those two species [2]. In 1960, Jane Goodall started studying wild chimpanzees in Tanzania, and Dian Fossey followed in 1967 in her study of mountain gorillas in Congo, and then Rwanda.

In 1971, encouraged by Dr. Louis Leakey, Birute Mary Galdikas began her field studies of orangutans in what is now known as Tanjung Puting National Park in Kalimantan Tengah, Indonesia. In many ways, orangutans are distinct from their great ape cousins. Orangutans are found in Asia instead of Africa and their hair is orange instead of black. But what made studying them especially challenging was the fact that they live solitary, mysterious lives high up in the trees instead of in large, obvious groups on the ground. After habituating the orangutans in her study area, Birute Mary Galdikas set out to study their behavior, and she has been following their individual life histories every since. Her studies reveal that, although orangutans spend far less time interacting with each other than humans, gorillas and chimpanzees do, orangutan social networks are intricate and form life-long bonds. Biruté Mary Galdikas likes to say that in contrast to the other great apes, orangutans never left the “Garden of Eden”. Orangutans remain primarily arboreal and semi-solitary. Warfare, which is common in human and chimpanzee societies, has never been observed among orangutans. The behavioral studies by Leakey’s Angels have yielded fascinating insights into the differences as well as similarities between humans and our closest living relatives, thereby providing an important piece to the puzzle of the evolution of great apes.
Today, studying life at the molecular level has become an indispensible tool that complements and often supports findings made through traditional methods such as archaeology (analysis of fossils), morphology (comparison of body part characteristics), and ethology (behavioral studies). To study evolution at the molecular level, researchers have in recent years sequenced the genomes of many different organisms. In all animals, plants, and bacteria, the genome consists of DNA, and it contains all the hereditary information necessary to keep the organism alive and functioning. Every single cell in our body contains a copy of the genome, giving the cell access to the “building plans” for the molecules of life. By comparing the DNA sequences of different organisms, it is possible to clarify their evolutionary relationships: the DNA sequences of two closely related organisms (such as orangutan and human) will show more similarities than the DNA sequences of two more distantly related organisms (such as orangutan and horse).
Over the past two years, the genomes of both orangutans and gorillas have been sequenced, following the human genome in 2001 and the chimpanzee genome in 2005 [1, 3, 4, 5]. Overall, comparison of the DNA sequences has confirmed the evolutionary relationships previously known: human genomic DNA is 99% identical with the DNA found in a chimpanzee [4]; gorillas come in second in this comparison, closely followed by orangutans, who share 97% of our DNA [1, 2]. In addition to independently confirming what was previously known, each genome contains surprises, leading to unexpected findings and exciting discoveries that could not have been made otherwise.
For the orangutan genome, one of the surprises is its amazing stability over time [1]. Since the orangutan genome has changed considerably less than the genome of any other primate, orangutans are likely much more closely related to the last common ancestor shared between all great apes than any other living species [1]. Therefore, among current life on earth, orangutans are the most similar to the ancestral great ape Louis Leakey wanted to find. This finding shook up a lot of entrenched notions, since it had previously been assumed that all primate genomes evolved at a similar pace [6]. If this had been the case, the genome of modern orangutans would be as similar to that of the ancestral great ape as the genomes of modern chimpanzees and humans are to it.
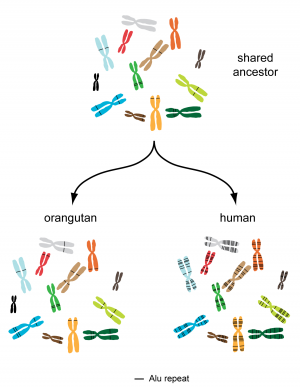
What may be the cause for the unusual stability of the orangutan genome? Analysis of the DNA indicates that the answer might lie in so-called “Alu elements” [1]. Alu elements are mobile genetic elements – in contrast to regular genes, Alu elements do not stay put in a specific location in the chromosome, but instead they duplicate and then get inserted into new positions. Thereby, the number of Alu elements in the genome steadily increases. Alu elements can therefore be thought of as “jumping genes”, and have been identified as one of the major driving forces of human evolution [7]. In the orangutan genome, Alu elements have been remarkably inactive: There have been only ~250 recent “jumps”, compared to 5,000 in humans and 2,300 in chimpanzees [1].
The unexpectedly low “jumping rate” of Alu elements is therefore the most likely explanation for the surprising stability of the orangutan genome. While the genomes of the other great apes have been constantly rearranged as a result of Alu elements “jumping” to new places, sometimes leading to dramatic reorganizations, the orangutan genome has evolved at a much slower pace. The close kinship at the molecular level between modern orangutans and the ancestor of all great apes is likely to be reflected at the macroscopic level as well, such as in their morphology and maybe even their behavior [8]. The sequence of the orangutan genome has provided an important piece to the puzzle that complements and extends what we know about the evolution of the great ape family from fossil, morphological and behavioral studies. Louis Leakey, if he were alive today, would probably have been thrilled and amazed to know that the elusive red ape, which had long been thought to be our most distant cousin, might hold such a critical clue to our ancient evolutionary history.
[1] Locke, D. P., Hillier, L. W., Warren, W. C. et al. (2011). Comparative and demographic analysis of orang–utan genomes. Nature, 469, 529–533.
[2] Virginia Morell (1995). Ancestral Passions. Touchstone publishing.
[3] Scally, A., Dutheil, J. Y., Hillier, L. W., et al. (2012). Insights into hominid evolution from the gorilla genome sequence. Nature, 483, 169–175.
[4] International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921.
[5] The Chimpanzee Sequencing and Analysis Consortium (2005). Initial sequence of the chimpanzee genome and comparison with the human genome. Nature, 437, 69–87.
[6] Easteal, S. (1991). The relative rate of DNA evolution in primates. Mol. Biol. Evol., 8 (1), 115–127.
[7] Kazazian, H. H. (2004). Mobile elements: Drivers of genome evolution. Science, 303, 1626–1632.
[8] Di Fiore, A. (2003). Molecular genetic approaches to the study of primate behavior, social organization, and reproduction. American Journal of Physical Anthropology, 122, 62–99.



